The CBMcarb-DB database contains the 3D structures of CBM binding polysaccharides and oligosaccharides. The following options for searching are available: Searching by keywords Searching by SCM Searching by fields Scrolling Through the whole databaseWhat is CBMcarb-DB ?
CBMcarb-DB is dedicated to display and analyse three-dimensional structures of carbohydrate binding modules (CBMs) in complex with carbohydrates, providing curated structural data about CBM carbohydrate complexes, binding interactions and carbohydrate conformation, with functional information on binding specificity.
What are CBMs ?CBMs can be defined as discrete auxiliary protein modules with a non-catalytic carbohydrate binding function. CBMs are typically associated to modular enzymes but can also be found isolated from a polypeptide chain or associated to other non-enzymatic proteins. Some lectins and LysM domains are also classified as CBMs. CBMs have been grouped into families based on amino acid sequence similarities. These modules show a highly diverse range of carbohydrate binding specificities and exert critical functions in enhancing enzyme catalytic efficiency (e.g. substrate targeting), as carbohydrate sensing domains or as adhesion molecules (e.g. mediating bacterial adhesion to host cell surface). CBMs are also used in different biotechnological applications.
PDB: PDB Code UniProt: UniProt Accession Code GlyToucan: GlyToucan Identification of the Glycan Organism: Origin of the protein or of GAG polysaccharide Function / Process / Cellular Compartiment : Biological role in compliance with Gene Ontology terms Resolution: Value in Angstrom of the crystal resolution. (set to 3 for fiber; to 0 otherwise) cbm_name: Name of the GAG cbm_max_length: Number of constituting monosaccharides Macromolecule-name: Protein or Polysaccharide or Virus name PDB_name: Title of the article as it appears in the PDB Method: Crystal Xray, Fiber Xray, NMR, Scattering, Modeling. PDB-ligand: Number and type of monosaccharide (3 letters PDB codes) cbm-mass: Molecular Mass of the GAG Pubmed: linked to publication Linucs: GAG nomenclature following (Linear Notation for Unique description of Carbohydrate Sequences For each structure a detailed page is available with 3D visualization, interactions and links to external databases see below.
Go to CBMcarb-DB homepage (https://cbmdb.glycopedia.eu/)
The database can be searched by keywords by textual input: i.e. organism, PDB code, UniProt accession number, protein name, CBM family, type of monosaccharide, oligosaccharide ligand, or textual fragments of the title of a publication:

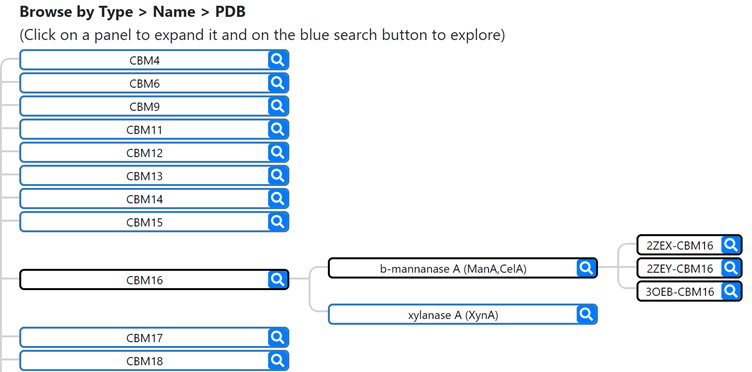
The search can be performed by the protein types included in each CBM family, by clicking on the desired panels to expand and acess each corresponding entry, or cliking in the blue search button to explore all the associated entries:
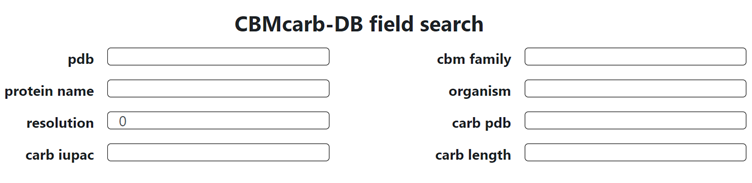
The database can be searched and explored with an advanced search tool handling a range of criteria: Pdb: PDB Code CBM Family: Family classification according to CAZy database Protein Name: Attributed name of the protein that includes the CBM module Organism: Origin of the protein Resolution: Value in Angstrom of the data resolution Carb PDB: Type of monosaccharide (3 letters PDB codes) Carb IUPAC: Oligosaccharide nomenclature according to the IUPAC condensed form Carb Length: Degree of polymerization of the oligosaccharide (For each structure, a detailed page is available with 3D visualization, interactions and links to external databases see below) On the homepage, click on the Easy Search for CBMs button: This will give access to the search by fields:
By clicking on any of the panels, it will display the existing content. The user is invited to select the entry of his/her choice or to search a desired content by typing on the corresponding panel, for example, typing a PDB ID in the pdb panel, a type of carbohydrate monomer such as “glc” in the carb iupac panel, or the degree of polymerization in the carb length panel. This is a multi-field selection process aiming at searching the structure and directing the user directly to access the desired target. This is achieved by clicking on the Scrolling through the database button: The selected criteria will lead to the corresponding entries:
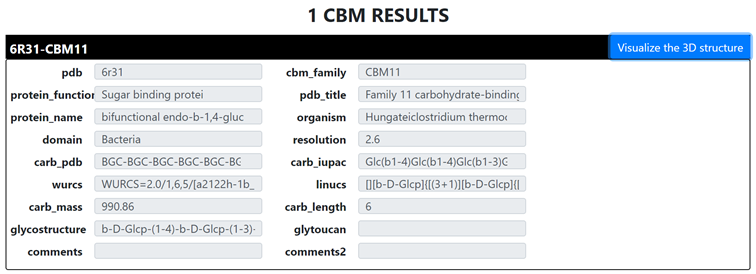
Clicking on 'visualize the 3D structure' yields to the detailed page:

In this page, an interactive view of the CBM-oligosaccharide is displayed. The LigPlot diagram, ligand sequence according to the SNFG symbol representation, the ligand and the CBM structures are also shown, and the user is able to download the respective images by clicking on them. For CBM complexes with ligands over 3 carbohydrate monomers, an interactive view of the oligosaccharide is also displayed, and the user can download the corresponding PDB file by clicking on the Oligo PDB file link. Links are also available to access other related information and resources: information about the original publication through PubMed or DOI; the protein sequence through UniProt, protein and carbohydrate structure and interaction using the various PDB databases, SwissModel, PLIP and PISA. Most of the structures in CBMcarb-DB were solved by X-ray crystallography methods, and in these cases an associated resolution value is shown in the corresponding result panel. When a structure was solved by NMR methods, “0” will appear in the resolution panel.
The content of the whole CBMcarb-DB can be visualized by scrolling through the entries displayed in the Field Search page:

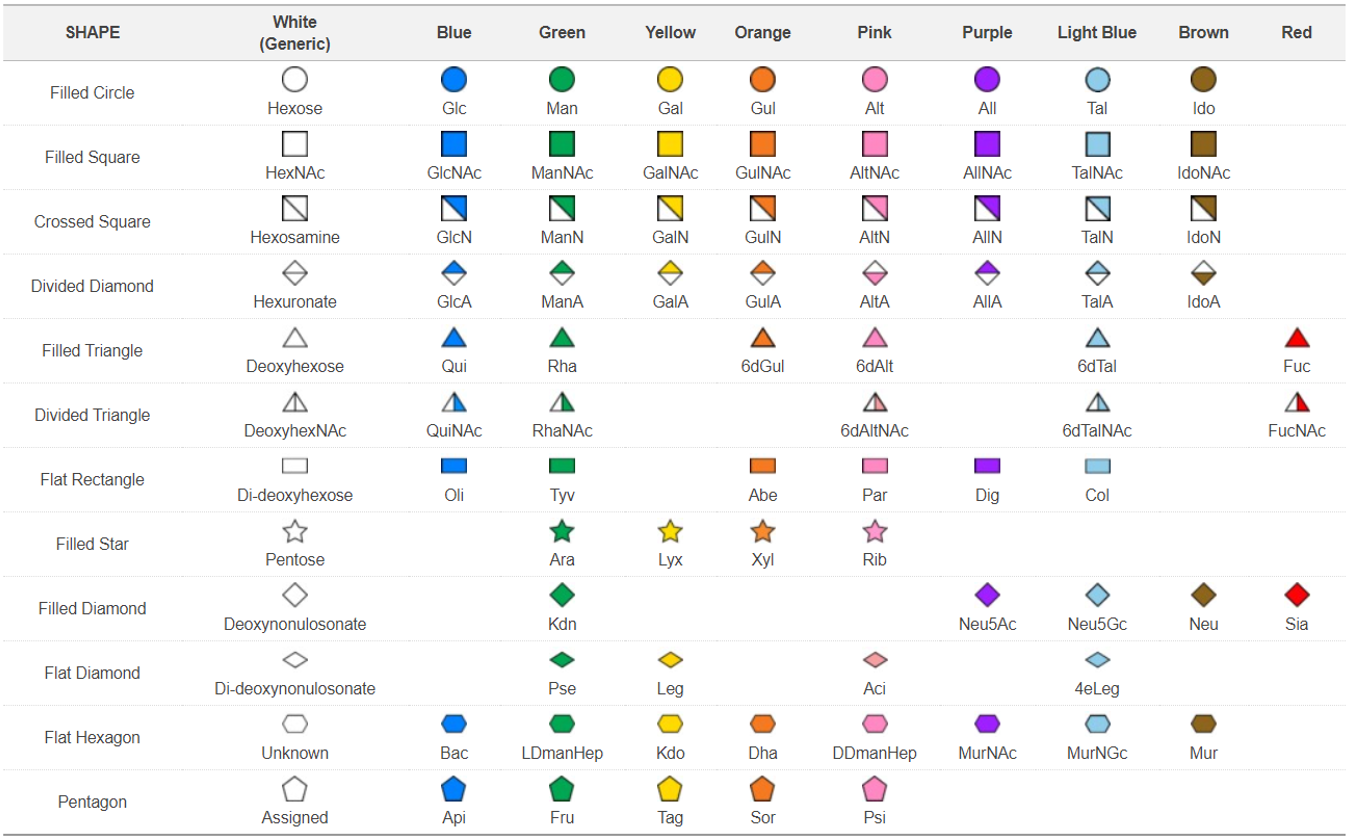
Third Edition of Essentials of Glycobiology - Appendix 1B: Symbol Nomenclature for Glycans (SNFG) Symbol Nomenclature for Graphical Representation of Glycans, Glycobiology 25: 1323-1324, 2015. Citation link (PMID 26543186).